Abstract
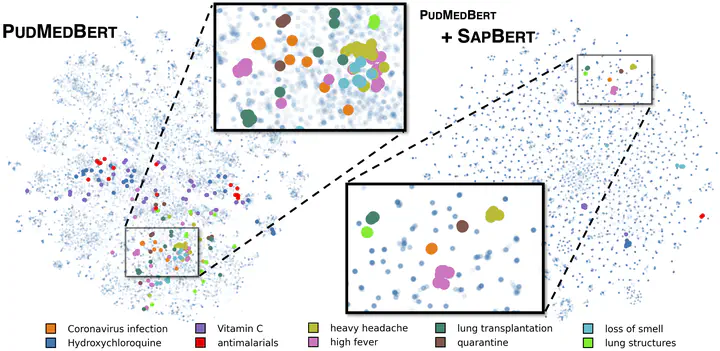
Despite the widespread success of self-supervised learning via masked language models (MLM), accurately capturing fine-grained semantic relationships in the biomedical domain remains a challenge. This is of paramount importance for entity-level tasks such as entity linking where the ability to model entity relations (especially synonymy) is pivotal. To address this challenge, we propose SapBERT, a pretraining scheme that self-aligns the representation space of biomedical entities. We design a scalable metric learning framework that can leverage UMLS, a massive collection of biomedical ontologies with 4M+ concepts. In contrast with previous pipeline-based hybrid systems, SapBERT offers an elegant one-model-for-all solution to the problem of medical entity linking (MEL), achieving a new state-of-the-art (SOTA) on six MEL benchmarking datasets. In the scientific domain, we achieve SOTA even without task-specific supervision. With substantial improvement over various domain-specific pretrained MLMs such as BioBERT, SciBERTand and PubMedBERT, our pretraining scheme proves to be both effective and robust.
Supplementary notes can be added here, including code, math, and images.