Biography
I currently hold the position of Lecturer (Assistant Professor) at the University of Glasgow, within the esteemed Information Retrieval Group and IDA section of the School of Computing Science. Additionally, I serve as an Affiliated Lecturer at the Language Technology Lab (LTL) at the University of Cambridge.
Previously, I conducted research as a Postdoctoral Researcher at the Language Technology Lab (LTL) of the University of Cambridge and as a Postdoctoral Researcher at the Information Retrieval group of the University of Glasgow. I have also been a visiting PhD student at the MINE lab of KAUST.
My research topics includes, Natural Language Processing, Information Retrieval, LLMs, AI Agents, as well as Knowledge (Graphs) Extraction, Representation & Reasoning Learning, particularly within BioMedical applications (AI4Biomedicine).
I co-lead the Glasgow AI4BioMed Lab, where we work on natural language processing, knowledge graphs, language models, and more to extract and infer biomedical knowledge. If you are interested in collaborating or pursuing a PhD with us, please refer to this post and this page for more details.
I will no longer be available to take on new PhD students this year.
- Natural Language Processing
- AI for BioMedicine
- Information Retrieval
- Knowledge Graph
- Geometric Deep Learning
- Recommender Systems
- Machine Learning
PhD in Computer Science, 2018
Sun Yat-sen University
M.Eng. in Computer Science, 2014
Guangdong University of Technology
B.Eng. in Software Engineering, 2010
Jiangxi University of Science and Technology
Diploma in Software Technology, 2008
Jiangxi University of Science and Technology (NanChang Campus)
News
- 2025-8-20: 🥳 Three papers were accepted by EMNLP 2025 on FusionDTI for drug-target interaction, Long-Tail Biomedical Knowledge Editing, and temporal reasoning evaluation, see you in Suzhou!
- 2025-8-12: 📢 Excited to announce our new survey paper: A Comprehensive Survey of Self-Evolving AI Agents: A New Paradigm Bridging Foundation Models and Lifelong Agentic Systems! 🌱🤖 A deep dive into techniques that bridge foundation models with lifelong agentic systems, featuring a unified framework, domain-specific strategies, and insights on evaluation, safety, and ethics. Read the full survey 👉 arXiv | Explore resources on GitHub
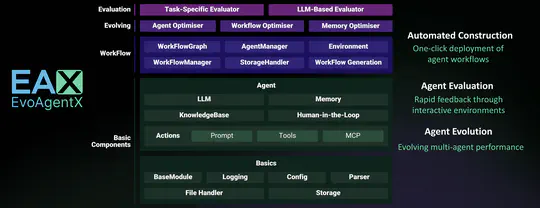
- 2025-5-15: 🚀 Thrilled to introduce EvoAgentX: The world’s first self-evolving framework for AI Agents!🌐 An automated framework for evaluating and evolving agentic workflows. Build and evolve your agents today at 👉 EvoAgentX
- 2025-5-15: Two papers were accepted by ACL 2025 on Knowledge-enhanced retrieval-augmented generation (RAG) models and Temporally-aware multimodal large language model tailored for chest X-ray report generation, see you in Vienna.
- 2024-11-26: Delivered a tutorial on “Integrating Knowledge Graphs and Large Language Models for Advancing Scientific Research” with Qiang Zhang and Jiaoyan Chen at Learning on Graphs Conference 2024. Recordings are avaiable at here.
- 2024-09-20: Two papers were accepted by EMNLP 2024 on RAG models with reasoning chains and position bias in large language models.
- 2024-06-10: Two papers have been accepted at Briefings in Bioinformatics, on Automatic Biochemical Pathway Prediction and Gene-disease Association Prediction.
- 2024-05-16: Two papers were accepted by ACL 2024 on Retrieval-Augmented Generation with Knowledge Graph Generation and Medical Open-Domain Question Answering.
- 2024-03-28: As the co-organization chair, we are organising the KEIR@ECIR 2024 workshop in Glasgow, UK.
- 2024-03-24: As the local organization chair, we are organising the ECIR 2024 conference in Glasgow, UK.
- 2024-01-16: Our paper entitled “CLEX: Continuous Length Extrapolation for Large Language Models” was accepted by ICLR 2024.
- 2023-12-17: Our paper entitled “BAND: Biomedical Alert News Dataset” was accepted by AAAI 2024.
- 2023-12-10: Invited to serve as one of the Area Chairs for NAACL 2024.
- 2023-10-9: Two papers were accepted by EMNLP 2023 on Unsupervised Biomedical NER and Multimodal Generative Language Model.
- 2023-09-26: Our survey paper on Knowledge Graph Embedding was accepted by ACM Computing Surveys.
- 2023-08-14: Our survey paper on Multimodal Language Modelling was accepted by ACM Transactions on Multimedia Computing, Communications, and Applications
- 2023-08-05: One paper was accepted by CIKM 2023 on Knowledge-enhance Passage Ranking.
- 2023-06-26: Invited to serve as one of the Area Chairs for the track “Interpretability, Interactivity and Analysis of Models for NLP” of EMNLP 2023.
- 2023-06-09: I gave an invited talk on “Probing and Infusing Biomedical Knowledge for Pre-trained Language Models” in “NLP for Social Good (NSG) Symposium 2023” hosted by Dr Procheta Sen at the University of Liverpool.
- 2023-05-22: One paper was accepted by Matching ACL 2023 on Generative Event Extraction.
- 2023-05-02: One paper was accepted by ACL 2023 on Few-shot NER.
- 2023-03-02: One paper was accepted by Transactions on the Web (TWEB) on Conversational Recommendation Systems.
- 2023-02-10: I gave a guest lecture on “Words Sense and WordNet” for the “LI18 - Computational Linguistics” course offered by Professor Nigel Collier at the University of Cambridge.
- 2023-01-24: I am an Area Chair for the “Interpretability and Analysis of Models for NLP” track of ACL 2023.
- 2023-01-21: One paper was accepted by EACL 2023 on Deductive Reasoning Analysis of Pretrained models. The preprint of it can be found in this link.
- 2022-10-25: I will be attending EMNLP 2022 (Abu Dhabi, UAE 🇦🇪) in person.
- 2022-10-06: One paper was accepted by EMNLP 2022 on Parameter-Efficient Tuning.
- 2022-10-04: Our paper entitled Graph Neural Pre-training for Recommendation with Side Information was accepted at ACM TOIS.
Projects
Featured Publications
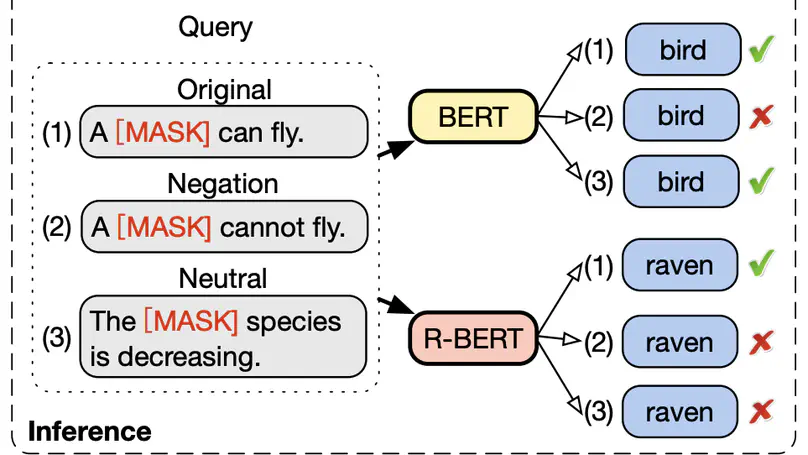
Acquiring factual knowledge with Pretrained Language Models (PLMs) has attracted increasing attention, showing promising performance in many knowledge-intensive tasks. Their good performance has led the community to believe that the models do possess a modicum of reasoning competence rather than merely memorising the knowledge. In this paper, we conduct a comprehensive evaluation of the learnable deductive (also known as explicit) reasoning capability of PLMs. Through a series of controlled experiments, we posit two main findings. (i) PLMs inadequately generalise learned logic rules and perform inconsistently against simple adversarial surface form edits. (ii) While the deductive reasoning fine-tuning of PLMs does improve their performance on reasoning over unseen knowledge facts, it results in catastrophically forgetting the previously learnt knowledge. Our main results suggest that PLMs cannot yet perform reliable deductive reasoning, demonstrating the importance of controlled examinations and probing of PLMs’ reasoning abilities; we reach beyond (misleading) task performance, revealing that PLMs are still far from human-level reasoning capabilities, even for simple deductive tasks.
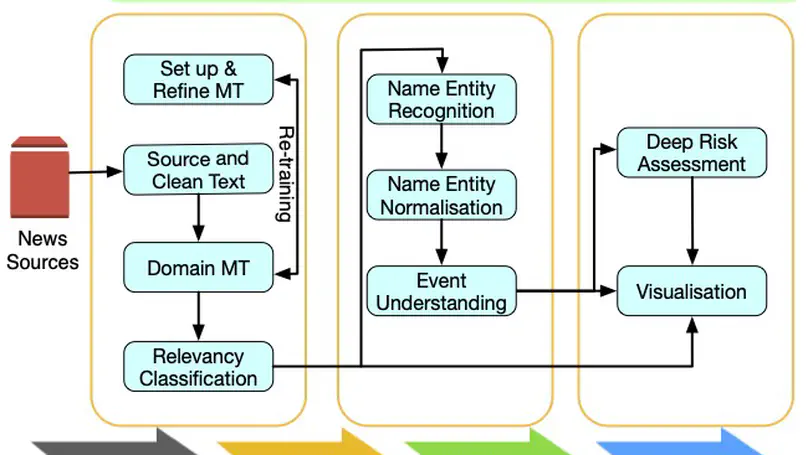
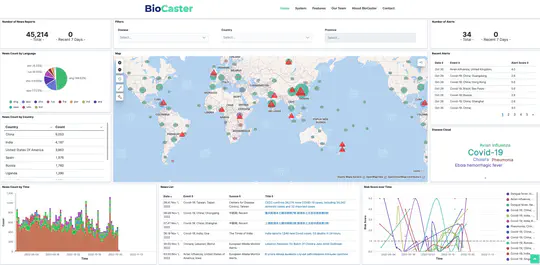
BioCaster was launched in 2008 to provide an ontology-based text mining system for early disease detection from open news sources. Following a 6-year break, we have re-launched the system in 2021. Our goal is to systematically upgrade the methodology using state-of-the-art neural network language models, whilst retaining the original benefits that the system provided in terms of logical reasoning and automated early detection of infectious disease outbreaks. Here, we present recent extensions such as neural machine translation in 10 languages, neural classification of disease outbreak reports and a new cloud-based visualization dashboard. Furthermore, we discuss our vision for further improvements, including combining risk assessment with event semantics and assessing the risk of outbreaks with multi-granularity. We hope that these efforts will benefit the global public health community.
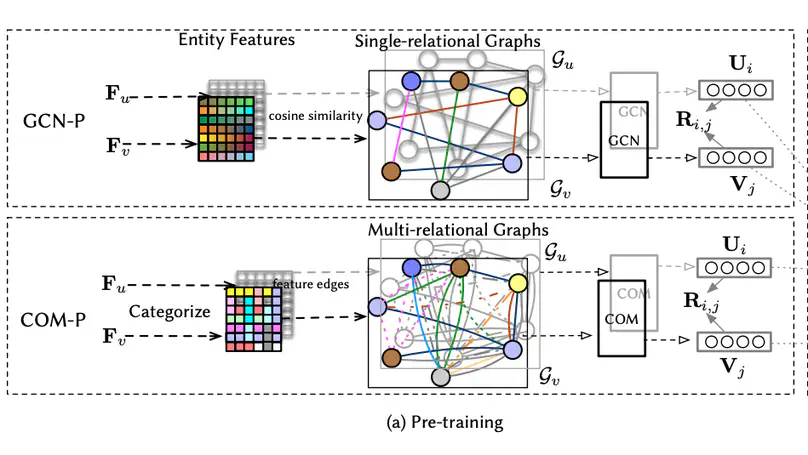
Leveraging the side information associated with entities (i.e. users and items) to enhance the performance of recommendation systems has been widely recognized as an important modelling dimension. While many existing approaches focus on the integration scheme to incorporate entity side information – by combining the recommendation loss function with an extra side information-aware loss – in this paper, we propose instead a novel pre-training scheme for leveraging the side information. In particular, we first pre-train a representation model using the side information of the entities, and then fine-tune it using an existing general representation-based recommendation model. Specifically, we propose two pre-training models, named GCN-P and COM-P, by considering the entities and their relations constructed from side information as two different types of graphs respectively, to pre-train entity embeddings. For the GCN-P model, two single-relational graphs are constructed from all the users’ and items’ side information respectively, to pre-train entity representations by using the Graph Convolutional Networks. For the COM-P model, two multi-relational graphs are constructed to pre-train the entity representations by using the Composition-based Graph Convolutional Networks. An extensive evaluation of our pre-training models fine-tuned under four general representation-based recommender models, i.e. MF, NCF, NGCF and LightGCN, shows that effectively pre-training embeddings with both the user’s and item’s side information can significantly improve these original models in terms of both effectiveness and stability.
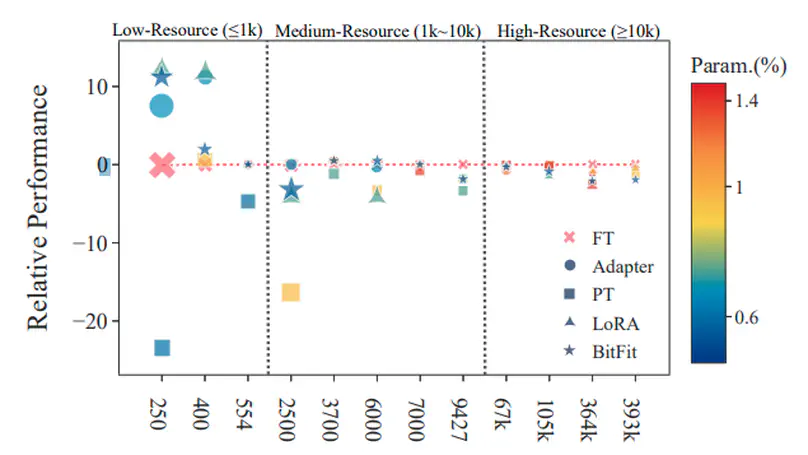
Parameter-efficient tuning (PETuning) methods have been deemed by many as the new paradigm for using pretrained language models (PLMs). By tuning just a fraction amount of parameters comparing to full model finetuning, PETuning methods claim to have achieved performance on par with or even better than finetuning. In this work, we take a step back and re-examine these PETuning methods by conducting the first comprehensive investigation into the training and evaluation of PETuning methods. We found the problematic validation and testing practice in current studies, when accompanied by the instability nature of PETuning methods, has led to unreliable conclusions. When being compared under a truly fair evaluation protocol, PETuning cannot yield consistently competitive performance while finetuning remains to be the best-performing method in medium- and high-resource settings. We delve deeper into the cause of the instability and observed that model size does not explain the phenomenon but training iteration positively correlates with the stability.
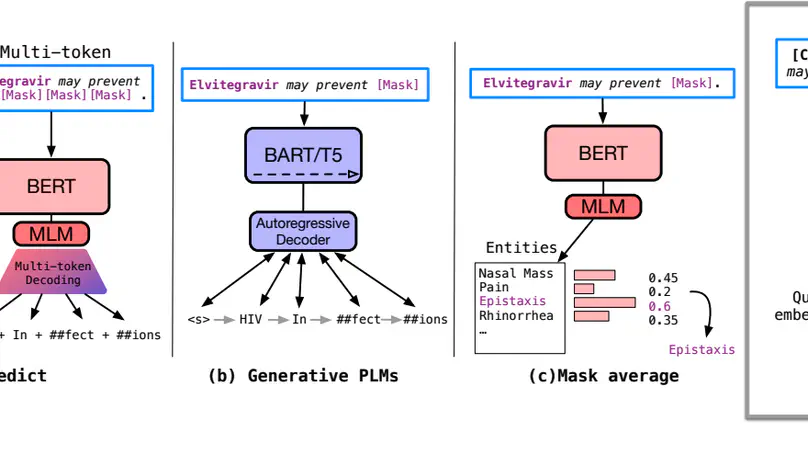
Knowledge probing is crucial for understanding the knowledge transfer mechanism behind the pre-trained language models (PLMs). Despite the growing progress of probing knowledge for PLMs in the general domain, specialised areas such as biomedical domain are vastly under-explored. To catalyse the research in this direction, we release a well-curated biomedical knowledge probing benchmark, MedLAMA, which is constructed based on the Unified Medical Language System (UMLS) Metathesaurus. We test a wide spectrum of state-of-the-art PLMs and probing approaches on our benchmark, reaching at most 3% of acc@10. While highlighting various sources of domain-specific challenges that amount to this underwhelming performance, we illustrate that the underlying PLMs have a higher potential for probing tasks. To achieve this, we propose Contrastive-Probe, a novel self-supervised contrastive probing approach, that adjusts the underlying PLMs without using any probing data. While Contrastive-Probe pushes the acc@10 to 28%, the performance gap still remains notable. Our human expert evaluation suggests that the probing performance of our Contrastive-Probe is still under-estimated as UMLS still does not include the full spectrum of factual knowledge. We hope MedLAMA and Contrastive-Probe facilitate further developments of more suited probing techniques for this domain.
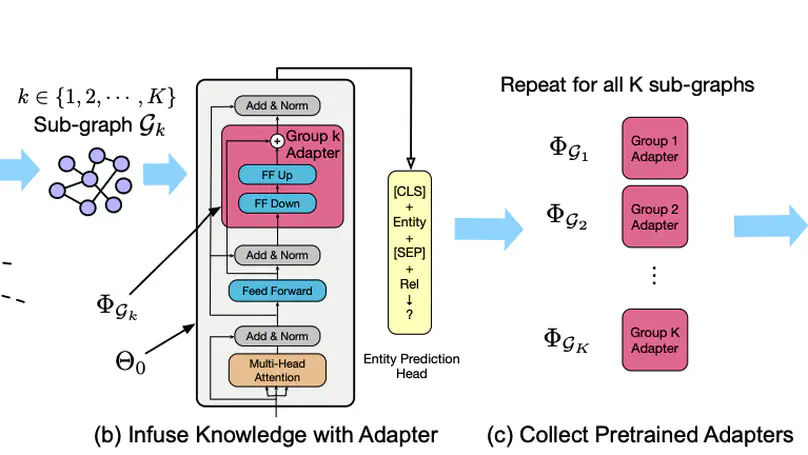
Infusing factual knowledge into pre-trained models is fundamental for many knowledge-intensive tasks. In this paper, we proposed Mixture-of-Partitions (MoP), an infusion approach that can handle a very large knowledge graph (KG) by partitioning it into smaller sub-graphs and infusing their specific knowledge into various BERT models using lightweight adapters. To leverage the overall factual knowledge for a target task, these sub-graph adapters are further fine-tuned along with the underlying BERT through a mixture layer. We evaluate our MoP with three biomedical BERTs (SciBERT, BioBERT, PubmedBERT) on six downstream tasks (inc. NLI, QA, Classification), and the results show that our MoP consistently enhances the underlying BERTs in task performance, and achieves new SOTA performances on five evaluated datasets.
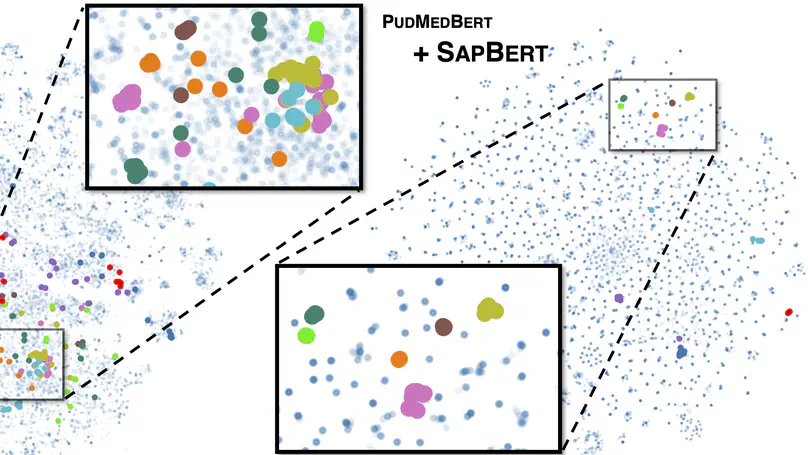
Despite the widespread success of self-supervised learning via masked language models (MLM), accurately capturing fine-grained semantic relationships in the biomedical domain remains a challenge. This is of paramount importance for entity-level tasks such as entity linking where the ability to model entity relations (especially synonymy) is pivotal. To address this challenge, we propose SapBERT, a pretraining scheme that self-aligns the representation space of biomedical entities. We design a scalable metric learning framework that can leverage UMLS, a massive collection of biomedical ontologies with 4M+ concepts. In contrast with previous pipeline-based hybrid systems, SapBERT offers an elegant one-model-for-all solution to the problem of medical entity linking (MEL), achieving a new state-of-the-art (SOTA) on six MEL benchmarking datasets. In the scientific domain, we achieve SOTA even without task-specific supervision. With substantial improvement over various domain-specific pretrained MLMs such as BioBERT, SciBERTand and PubMedBERT, our pretraining scheme proves to be both effective and robust.
Professional Service
Conference/Workshop Organisers:
- The European Conference on Information Retrieval (ECIR) 2024, Local Organisation Chair
- The First Workshop on Knowledge-Enhanced Information Retrieval workshop (KEIR @ ECIR 2024), Workshop Chair
Conference Area Chair/Senior Program Committee:
- The Annual Meeting of the Association for Computational Linguistics (ACL), 2023
- The Conference on Empirical Methods in Natural Language Processing (EMNLP), 2023
Conference Program Committee/Reviewer:
- Conference On Language Modeling (COLM), 2024
- International Conference on Machine Learning (ICML), 2021, 2022, 2023, 2024
- The Conference on Neural Information Processing Systems (NeurIPS), 2020, 2021, 2022, 2023, 2024
- The International Conference on Learning Representations (ICLR), 2021, 2022, 2023, 2024
- The Annual Meeting of the Association for Computational Linguistics (ACL), 2022, 2023, 2024
- ACL Rolling Review, 2021, 2022, 2023, 2024
- The Conference on Empirical Methods in Natural Language Processing (EMNLP), 2018, 2021, 2022
- The 17th Conference of the European Chapter of the Association for Computational Linguistics (EACL), 2023
- The ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR), 2018, 2021, 2022, 2023, 2024
- The Conference on Information and Knowledge Management (CIKM), 2017, 2018
- The ACM Conference on Web Search and Data Mining (WSDM), 2018, 2019
- The European Conference on Information Retrieval (ECIR), 2020, 2021, 2022, 2023, 2024
- The ACM Web Conference (WWW), 2018, 2021, 2022, 2023,2024
- The ACM SIGKDD Conference on Knowledge Discovery and Data Mining (SIGKDD), 2021, 2022
- The AAAI Conference on Artificial Intelligence (AAAI), 2020, 2021, 2022, 2023, 2024
- The ACM Recommender Systems Conference (RecSys), 2020, 2021, 2022
- International Joint Conference on Artificial Intelligence (IJCAI), 2018, 2020, 2022
- The SIAM International Conference on Data Mining (SDM), 2022
Journal Editor/Guest Editor:
- Special Issue of Electronics (ISSN 2079-9292) journal on “Natural Language Processing and Information Retrieval”.
Journal Invited Reviewer:
- ACM Transactions on Information Systems
- ACM Transactions on the Web
- ACM Computing Surveys
- Information Retrieval Journal
- ACM Transactions on Recommender Systems
- Computers in Biology and Medicine
- IEEE Transactions on Knowledge and Data Engineering
- IEEE Transactions on Neural Networks and Learning Systems
- IEEE Transactions on Cybernetics
- Information Sciences
- World Wide Web
- IEEE Transactions on Fuzzy Systems
- IEEE Access
- International Journal of Machine Learning and Cybernetics
- Concurrency and Computation: Practice and Experience
Teaching
Contact
- zaiqiao.meng@glasgow.ac.uk
- 220b Sir Alwyn Williams Building, Glasgow, UK, G128QQ
-
Monday 09:00 to 18:00
Friday 09:00 to 18:00 - Follow Me